- Standalone Applications
- BioEddie
- Chemaxon Assay
- ChemCurator
- ChemLocator
- cHemTS
- Compliance Checker
- Compound Registration
- Quick Start Guide
- Compound Registration User's Guide
- Overview
- Compound Registration Abbreviations
- Definitions of Terms
- Compound Registration Introduction
- Login
- Dashboard page
- Autoregistration
- Bulk Upload
- Advanced Registration
- Search
- User Profile
- Browse page
- Appendix A. Calculations
- Appendix B. Markush Structures
- Multi-Component compounds
- Restricted compounds
- Configuration Guide
- Deployment Guide
- Compound Registration FAQ
- Compound Registration System Requirements
- Compound Registration History of Changes
- API documentation
- Instant JChem
- Instant Jchem User Guide
- Getting Started
- IJC Projects
- IJC Schemas
- Viewing and Managing Data
- Lists and Queries
- Collaboration
- Import and Export
- Editing Databases
- Relational Data
- Chemical Calculations and Predictions
- Chemistry Functions
- Security
- Scripting
- Updating Instant JChem
- Tips and Tricks
- Instant JChem Tutorials
- Building a relational form from scratch
- Building more complex relational data models
- Defining a security policy
- Filtering items using roles
- Lists and Queries management
- Query building tutorial
- Reaction enumeration analysis and visualization
- SD file import basic visualization and overlap analysis
- Using Import map and merge
- Using Standardizer to your advantage
- Pivoting tutorial
- Instant JChem Administrator Guide
- Admin Tool
- IJC Deployment Guide
- Supported databases
- JChem Cartridge
- Using Oracle Text in Instant JChem
- Deployment via Java Web Start
- Startup Options
- Shared project configuration
- Accessing data with URLs
- Instant JChem Meta Data Tables
- Test to Production Metadata Migrator
- Filtering Items
- Deploying the IJC OData extension into Spotfire
- Reporting a Problem
- Manual Instant JChem schema admin functions
- SQL Scripts for Manual Schema Upgrade
- Database Row Level Security
- JccWithIJC
- Instant JChem Developer Guide
- Working With IJC Architecture
- IJC API
- Groovy Scripting
- Good Practices
- Schema and DataTree Scripts
- Simple SDF Exporter
- Relational SDF Exporter
- CDX File Importer
- Data Merger or Inserter from an SDF file
- Markush DCR Structures Exporter
- Select Representative Member of Clusters
- Table Standardizer
- Populate a Table with Microspecies
- Create a Diverse Subset
- Pearson Linear Correlation Co-efficient Calculator
- PDF Trawler
- Simple Substructure Search
- Intersecting Sets
- Find Entries with Duplicated Field Value
- Importing Multiple SDF Files
- Calling External Tools
- Create Relational Data Tree
- Forms Model Scripts
- Button Scripts
- Execute Permanent Query
- Patent Fetcher Button
- Batch Searching Button
- Import or Export a Saved Query SDF Button
- Back and Next Buttons
- Add Annotations Button
- Simple Structure Checker Button
- Advanced Structure Checker Button
- Calculate MolWeight and generate SMILES
- Get Current User
- Simple ChemicalTerms evaluator
- Edit Molecule Button
- TanimotoMultiple
- Execute Permanent Query Based On Its Name
- Open existing view in the same dataTree
- Export selection to file
- Generate random resultset from actual resultset
- Form Scripts
- Groovy Scriptlets
- Buttons vs Scripts
- Creating New Entities
- Creating New Fields
- Reading Molecules From a File
- Insert or Update a Row
- Evaluator
- Create or Find a Relationship
- Adding an Edge to a Data Tree
- Exporting Data to a File
- Connect to an External Database
- Create a New ChemTerm Field
- Create a New Dynamic URL Field
- Create a New Static URL Field
- Java Plugins
- IJC Plugin Quick Start
- IJC Hello World Plugin
- IJC Plugin tutorial - MyAddField plugin
- IJC Plugin tutorial - MyMathCalc plugin
- IJC Plugin tutorial - Renderer Example
- IJC Plugin tutorial - MySCServer webapp
- IJC Plugin tutorial - MySCClient plugin
- IJC Plugin tutorial - Canvas widget
- Java Plugins and Java Web Start
- Instant JChem FAQ
- Instant JChem Installation and Upgrade
- New Features
- New Features in IJC Q4 2020
- New Features in IJC Q3 2020
- New Features in IJC Q2 2020
- New Features in IJC Q1 2020
- New Features in IJC Q4 2019
- New Features in IJC Q3 2019
- New Features in IJC Q2 2019
- New Features in IJC Q1 2019
- New Features in IJC Q4 2018
- New Features in IJC Q3 2018
- New Features in IJC Q2 2018
- New Features in IJC Q1 2018
- New Features in IJC Q4 2017
- New Features in IJC Q3 2017
- New Features in IJC Q2 2017
- New Features in IJC Q1 2017
- New Features in IJC Q4 2016
- New Features in IJC Q3 2016
- New Features in IJC Q2 2016
- New Features in IJC Q1 2016
- New Features in IJC Q4 2015
- New Features in IJC Q3 2015
- New Features in IJC Q2 2015
- New Features in IJC Q1 2015
- Instant JChem Licensing
- IJC Getting Help and Support
- Instant JChem System Requirements
- Instant JChem History of Changes
- Instant Jchem User Guide
- Markush Editor
- Design Hub
- History of changes
- Install guide
- Configuration guide
- Plugin Catalogue
- Developer guide - REST API
- Developer guide - resolver plugins
- Developer guide - real time plugins
- Developer guide - real time plugin templates
- Developer guide - export plugins
- Developer guide - storage plugins
- Developer guide - company support plugins
- Developer guide - registry plugins
- Developer guide - theme customization
- Install guide - Kubernetes
- Marvin Desktop Suite
- MarvinSketch
- User Guide
- Getting Started
- Graphical User Interface
- Working in MarvinSketch
- Structure Display Options
- Basic Editing
- Drawing Simple Structures
- Drawing More Complex Structures
- Drawing Reactions
- Using Integrated Calculations
- Graphical Objects
- Import and Export Options
- Multipage Documents
- Printing
- Chemical Features
- Marvin OLE User Guide
- Appendix
- Tutorials
- Developer Guide
- MarvinSketch Application Options
- User Guide
- MarvinView
- Marvin Desktop Suite Installation and Upgrade
- Marvin Desktop Suite History of Changes
- Marvin Desktop Suite Licensing
- MarvinSketch
- Molconvert
- Plexus Connect
- Plexus Connect - Quick Start Guide
- Plexus Connect - User Guide
- Plexus Connect - Log in
- Plexus Connect - Dashboard
- Plexus Connect - Exporting Your Data
- Plexus Connect - Export Templates
- Plexus Connect - Browsing in Your Data Set
- Plexus Connect - Selecting Data
- Plexus Connect - Searching in Your Database
- Plexus Connect - Saved Queries
- Plexus Connect - List Management
- Plexus Connect - Sorting Data
- Plexus Connect - Sharing Data with Other Users
- Plexus Connect - Charts View
- Plexus Connect - R-group Decomposition
- Plexus Connect - Administrator Guide
- Plexus Connect - Authentication
- Plexus Connect - Sharing Schema Items Among Users
- Plexus Connect - Business Flags
- Plexus Connect - Row-level Security
- Plexus Connect - Shared data sources
- Plexus Connect - Plexus storage
- Plexus Connect - Configuration Files
- Plexus Connect - Simple table
- Plexus Connect - Getting the Plexus Backend and Frontend Log Files
- Plexus Connect - Form Editor
- Plexus Connect - Scripting
- Plexus Connect - API keys
- Plexus Connect - Deploying Spotfire Middle Tier solution
- Plexus Connect - Installation and System Requirements
- Plexus Connect - Licensing
- Plexus Connect - Getting Help and Support
- Plexus Connect - FAQ
- Plexus Connect - Privacy Policy
- Plexus Connect - Terms of Use for the Demo Site
- Plexus Connect - History of Changes
- Plexus Connect - Schema Refresh Without Restart
- Zosimos
- Trainer Engine
- Toolkits and Components
- AutoMapper
- Calculator Plugins
- Introduction to Calculator Plugins
- Calculator Plugins User's Guide
- Calculator Plugins Developer's Guide
- Calculators on AWS Marketplace
- Background materials
- Calculation of partial charge distribution
- Generate3D
- Isoelectric point (pI) calculation
- LogP and logD calculations
- NMR model prediction
- pKa calculation
- Red and blue representation of pKa values
- Tautomerization and tautomers
- Validation results
- Tautomerization and tautomer models of Chemaxon
- Theory of aqueous solubility prediction
- The tautomerization models behind the JChem tautomer search
- Calculators performance reports
- Calculator Plugins Licensing
- Calculator Plugins FAQ
- Calculator Plugins Getting Help and Support
- Calculator Plugins History of Changes
- Calculator Plugins System Requirements
- Biomolecule Toolkit
- Chemaxon Synergy
- Chemaxon Synergy User Guide
- Chemaxon Synergy Developer Guide
- Chemaxon Synergy Administrator Guide
- Chemaxon Synergy History of Changes
- Document to Structure
- JChem Base
- JChem Base Administration
- JChem Base Developer's Guide
- JChem Base User's Guide
- Query Guide
- Search types
- Similarity search
- Query features JCB
- Stereochemistry JCB
- Special search types
- Search options
- Atomproperty specific search options
- Attached data specific search options
- Bond specific search options
- Chemical terms specific search options
- Database specific search options
- General search options
- Hitdisplay specific search options
- Markush structure specific search options
- Performance specific search options
- Polymer specific search options
- Query feature specific search options
- Reaction specific search options
- Resultset specific search options
- Similarity specific search options
- Stereo specific search options
- Tautomer specific search options
- Tautomer search - Vague bond search - sp-Hybridization
- Standardization JCB
- Hit display-coloring
- Appendix JCB
- Matching Query - Target Examples
- jcsearch Command Line Tool
- jcunique Command Line Tool
- Homology Groups in Markush Structures
- Query Guide
- JChem Base FAQ
- JChem Base History of Changes
- JChem Base Getting Help and Support
- JChem Choral
- JChem Oracle Cartridge
- JChem Microservices
- JChem PostgreSQL Cartridge
- Getting Started JChem PostgreSQL Cartridge
- JChem PostgreSQL Cartridge Manual
- Second Generation Search Engine
- JChem PostgreSQL Cartridge FAQ and Known Issues
- JChem PostgreSQL Cartridge History of Changes
- JChem PostgreSQL Cartridge Getting Help and Support
- JChem PostgreSQL Cartridge Licensing
- Comparison of JChem PostgreSQL Cartridge and JChem Oracle Cartridge
- Migration Guide
- JKlustor
- Markush Tools
- Marvin JS
- User Guide
- Getting Started
- Editor Overview
- Editor Canvas
- Dialogs
- Toolbars
- Context Menus
- Drawing and Editing Options
- Feature Overview Pages
- Keyboard Shortcuts
- Developer Resources
- History of Changes
- Frequently Asked Questions
- Video Tutorials
- Comparison of Marvin JS and MarvinSketch Feature Sets
- User Guide
- Marvin Pro
- Name to Structure
- Reactor
- Reactor User's Guide
- Introduction to Reactor
- Reactor Getting Started
- Reactor Concepts
- Reactor Examples
- Working with Reactor
- Specifying Reactions
- Specifying Reactants
- Reaction Mapping
- Reaction Rules
- Reactant Combinations
- Running Reactor
- Reactor Interfaces
- Reactor Application
- Reactor Command-line Application
- Reactor in Instant JChem
- Reactor in JChem for Excel
- Reactor in KNIME
- Reactor in Pipeline Pilot
- Reactor in Plexus Suite
- API, Web Services
- Glossary
- Reactor FAQ
- Reactor Licensing
- Reactor Getting Help and Support
- Reactor History of Changes
- Reactor Configuration Files
- Reactor User's Guide
- Screen
- Standardizer
- Standardizer User's Guide
- Standardizer Introduction
- Standardizer Getting Started
- Standardizer Concepts
- Working with Standardizer
- Standardizer Actions
- Add Explicit Hydrogens
- Alias to Atom
- Alias to Group
- Aromatize
- Clean 2D
- Clean 3D
- Clear Isotopes
- Clear Stereo
- Contract S-groups
- Convert Double Bonds
- Convert Pi-metal Bonds
- Convert to Enhanced Stereo
- Create Group
- Dearomatize
- Disconnect Metal Atoms
- Expand S-groups
- Expand Stoichiometry
- Map
- Map Reaction
- Mesomerize
- Neutralize
- Remove Absolute Stereo
- Remove Atom Values
- Remove Attached Data
- Remove Explicit Hydrogens
- Remove Fragment
- Remove R-group Definitions
- Remove Stereo Care Box
- Replace Atoms
- Set Absolute Stereo
- Set Hydrogen Isotope Symbol
- Strip Salts
- Tautomerize
- Transform
- Ungroup S-groups
- Unmap
- Wedge Clean
- Remove
- Standardizer Transform
- Custom Standardizer Actions
- Remove Solvents
- Creating a Configuration Standardizer
- Interfaces Standardizer
- Standardizer File Formats
- Standardizer Actions
- Standardizer Developer's Guide
- Standardizer Installation and System Requirements
- Standardizer Licensing
- Standardizer Getting Help and Support
- Standardizer History of Changes
- Standardizer User's Guide
- Structure Checker
- Structure Checker User's Guide
- Introduction
- Structure Checker Getting Started
- Structure Checker Concepts
- Working with Structure Checker
- Checker List
- Abbreviated Group StrCh
- Absent Chiral Flag
- Absolute Stereo Configuration
- Alias
- Aromaticity Error
- Atom Map
- Atom Query Property
- Atom Value
- Atropisomer
- Attached Data StrCh
- Bond Angle
- Bond Length
- Brackets
- Chiral Flag
- Chiral Flag Error
- Circular R-group Reference
- Coordination System Error
- Covalent Counterion
- Crossed Double Bond
- Custom Checkers and Fixers
- Double Bond Stereo Error
- EZ Double Bond
- Empty Structure
- Explicit Hydrogen
- Explicit Lone Pairs
- Incorrect Tetrahedral Stereo
- Isotope
- Metallocene Error
- Missing Atom Map
- Missing R-group Reference
- Molecule Charge
- Multicenter
- Multicomponent
- Multiple Stereocenter
- Non-standard Wedge Scheme
- Non-stereo Wedge Bond
- OCR Error
- Overlapping Atoms
- Overlapping Bonds
- Pseudo Atom
- Query Atom
- Query Bond
- Racemate
- Radical
- Rare Element
- R-atom
- Reacting Center Bond Mark
- Reaction Map Error
- Relative Stereo
- R-group Attachment Error
- R-group Bridge Error
- R-group Reference Error
- Ring Strain Error
- Solvent
- Star Atom
- Stereo Care Box
- Stereo Inversion Retention Mark
- Straight Double Bond
- Substructure
- Three Dimension 3D
- Unbalanced Reaction
- Unused R-group Reference
- Valence Error
- Valence Property
- Wedge Error
- Wiggly Bond
- Wiggly Double Bond
- Creating a Configuration StrCh
- Interfaces StrCh
- Checker List
- Structure Checker Developer's Guide
- Structure Checker Installation and System Requirements
- Structure Checker Licensing
- Structure Checker Getting Help and Support
- Structure Checker History of Changes
- Structure Checker User's Guide
- Structure to Name
- Third-party Integration
- Cross Product Documentation
- Chemaxon Configuration Folder
- Chemical Fingerprints
- Chemical Terms
- File Formats
- Basic export options
- Compression and Encoding
- Document formats
- Graphics Formats
- Molecule file conversion with Molconverter
- Molecule Formats
- CML
- MDL MOL files
- Daylight SMILES related formats
- Chemaxon SMILES extensions
- IUPAC InChI, InChIKey, RInChI and RInChIKey
- Name
- Sequences - peptide, DNA, RNA
- FASTA file format
- Protein Data Bank (PDB) file format
- Tripos SYBYL MOL and MOL2 formats
- XYZ format
- Gaussian related file formats
- Markush DARC format - VMN
- CSV
- Input and Output System
- License Management
- Long Term Supported Releases - LTS
- Notice about CAS Registry Numbers®
- Public Repository
- Scientific Background
- Structure Representation
- Structure Representation - Class Representation
- Aromaticity
- Implicit, Explicit and Query Hydrogens
- Assigning stereochemistry descriptors
- Cleaning options
- Deprecated and Removed Methods
- Relative configuration of tetrahedral stereo centers
- Iterator Factory
- Atom and bond-set handling
- Graphic object handling
- Supported Java Versions
- Legal
- Legal Documents
- Cookie Policy
- End User License Agreement
- End User Subscription Agreement - EUSA
- Personal Data Management - General
- Personal Data Management - Newsletter Subscriptions
- Privacy Policy
- Privacy Policy for Chemaxon Synergy
- Terms of Use
- Terms and Conditions for UX Research Program
- Notice for Chemaxon Software User Experience (UX) Research
- Recording Policy
- White Papers
- Legal Documents
- Discontinued Products
- Document to Database
- Fragmenter
- JChem Neo4j Cartridge
- Markush Overlap
- MarvinSpace
- MarvinSpace User's Guide
- MarvinSpace Developer's Guide
- MarvinSpace History of Changes
- Metabolizer
- Plexus Suite
- Quick Start Guide - Plexus Suite
- Plexus Suite User Guide
- Log in to Plexus Suite
- The Plexus Suite Dashboard
- Importing New Data
- Exporting Your Data
- Browsing in Your Data Set
- Selecting Data
- Searching in Your Database
- Saved Queries
- List Management
- Sorting Data
- Sharing Data with Other Users
- Calculating Molecular Properties for Single Compounds
- Adding calculated columns to tables
- Scaffold Based Enumeration
- Reaction Based Enumeration
- Registering Molecules in the Corporate Database
- Charts view
- Plexus Suite Video Tutorials
- Plexus Suite Administrator Guide
- Plexus Suite Authentication
- Sharing Schema Items Among Users
- Business Flags
- Row-level Security
- Building blocks
- Admin Tools
- Adding JavaScript Files for Custom Functionality
- Writing JavaScript Files for Custom Functionality
- Integration with Chemaxon's Compound Registration System
- Shared data sources in Plexus Connect
- Configuration Files
- Edit Views
- Simple table
- Installation and System Requirements of Plexus Suite
- Plexus Suite Licensing
- Getting Help and Support for Plexus Suite
- Plexus Suite FAQ
- Plexus Suite Privacy Policy
- Terms of Use for the Plexus Suite Demo Site
- Plexus Suite History of Changes
- Schema Refresh Without Restart
- JChem Web Services Classic
Geometrical Descriptors Plugin
This manual gives you a walk-through on how to use the Geometrical Descriptors Plugin:
Introduction
The Geometrical Descriptors Plugin calculates 3D geometric descriptors for a molecule. The following window shows a calculation result.
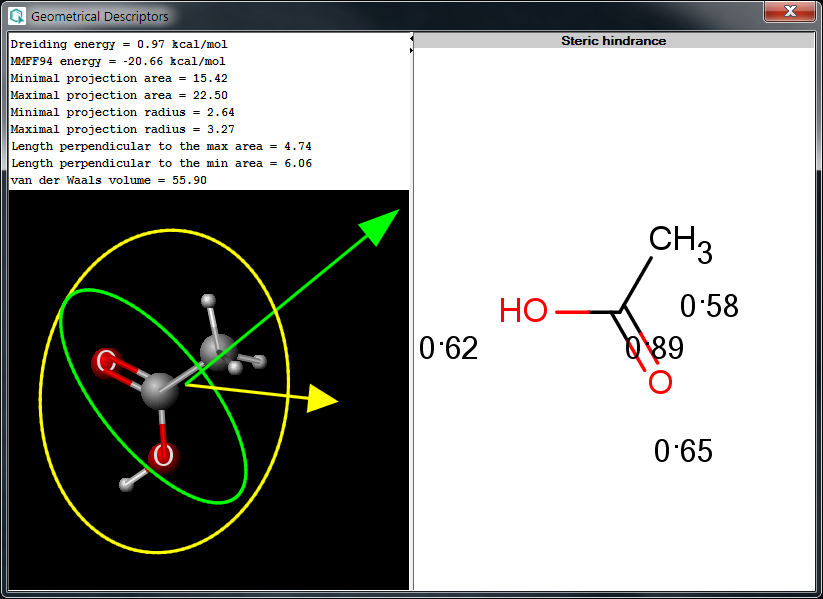
Fig.1 Geometrical Descriptors Plugin result window
Options
The following options can be set in the Geometrical Descriptors Options window:
-
Type
-
Dreiding energy: calculates the energy of the 3D structure (conformation) of the molecule using Dreiding force field.
-
MMFF94 energy: calculates the energy of the 3D structure (conformation) of the molecule using MMFF94 force field.
-
Steric hindrance: calculates steric hindrance of an atom calculated from the covalent radii values and geometrical distances.
-
Minimal projection area: calculates the minimum of projection areas of the conformer, based on the van der Waals radius (in Å2).
-
Maximal projection area: calculates the maximum of projection areas of the conformer, based on the van der Waals radius (in Å2).
-
Minimal projection radius: calculates the radius for the minimal projection area of the conformer (in Å).
-
Maximal projection radius: calculates the radius for the maximal projection area of the conformer (in Å).
-
Maximal distance perpendicular to the min projection: calculates the maximal extension of the conformer perpendicular to the minimal projection area (in Å).
-
Maximal distance perpendicular to the max projection: calculates the maximal extension of the conformer perpendicular to the maximal projection area (in Å).
-
van der Waals volume: calculates the van der Waals volume of the conformer (in Å3).
-
-
Energy unit: unit of the calculated energy (kcal/mol or kJ/mol).
-
Decimal places: setting the number of decimal places with which the result value is given.
-
Set MMFF94 optimalization : The structure is optimized before MMFF94 energy calculation.
-
Set projection optimalization The structure is optimized before projection area and projection radius calculation(s).
-
Calculate for lowest energy conformer:
-
If molecule is in 2D : the lowest energy conformer is generated if the input molecule is in 2D; 3D input molecules are considered in the given conformation.
-
Never : the input molecule is used for calculation.
-
Always : the lowest energy conformer is generated (for 3D and 2D molecules as well).
-
-
Optimization limit: sets the rigour of convergence for optimization. Stricter convergence means lower gradient limit to reach during optimization. The values can be:
-
Very loose
-
Normal
-
Strict
-
Very strict
-
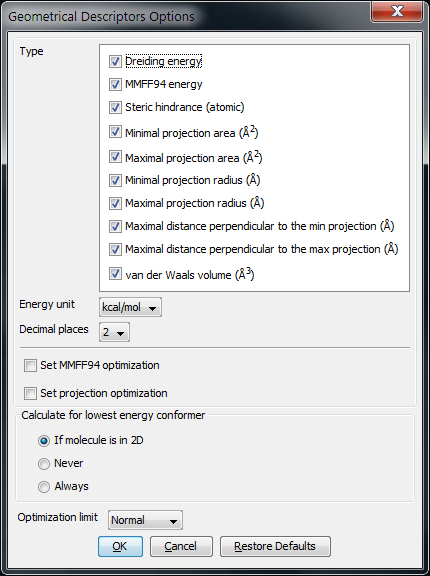
Fig. 2 Geometrical Descriptors Options window