CIP Stereo chemistry
Definitions
Tetrahedral stereo descriptor
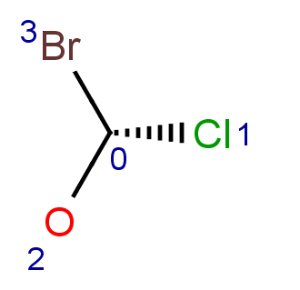
Parity is a simplified and local description of a chiral center. It is used for string the three dimensional arrangement of the ligands around a chiral center in a local canonical way without using the Cahn-Ingold-Prelog priority rules. The ligands of the chiral center are numbered according to the atom indexes in the molecule. If the atom has implicit Hydrogen, then this implicit Hydrogen atom is considered to be the highest numbered atom. The tetrahedral center atom is viewed such a way that the highest-numbered atom is behind the plane formed by the other three atoms. The three atoms can be arranged in either a clockwise or counterclockwise direction in ascending numerical order.
 |
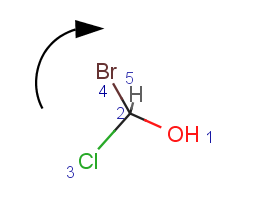 |
|---|---|
TetrahedralStereoValue.ODD |
Parity value TetrahedralStereoValue.ODD is assigned for a clockwise arrangement at the stereo center and TetrahedralStereoValue.EVEN for counterclockwise. If the center has wiggly (wavy) bond then TetrahedralStereoValue.WIGGLY value is assigned, otherwise if the given information is not enough to decide the spatial arrangement (e.g. no wedges in two dimensions) TetrahedralStereoValue.UNKNOWN value is assigned.
On this page
-


TetrahedralStereoValue.WIGGLYTetrahedralStereoValue.UNKNOWN
Atrop stereo descriptor
Hindered rotation around single bonds where the steric strain barrier to rotation is high enough to allow the isolation of the conformers resulting in atrop stereoisomerism.
Atrop stereo descriptor represents the topology of the case when two aromatic rings connected by a single bond with at least three ortho ligands.
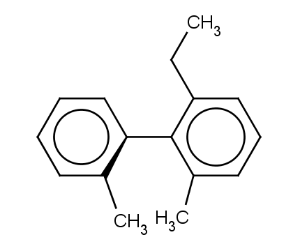
We look at the molecule in the direction of the single bond between the two aromatic rings (axis). On the example shown below the red arrow indicates the direction of the view.
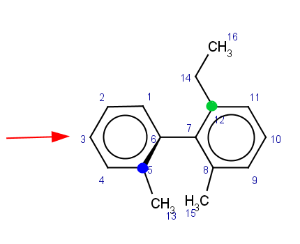
The axis is looked at from the direction where the highest indexed ligand connected to the axis (atom index 12) is behind the axis. The angle between two bonds connecting the highest indexed ligands on both sides to the axis is considered. On the example these are the bond between atoms 5 (marked blue) and 6 (in front of the axis) and the bond between atoms 12 (marked green) and 7.
-
AtropStereoValue.EVENmeans that the angle is positive (between 0 and Pi). -
AtropStereoValue.ODDmeans that the angle is negative (between 0 and -Pi). -
AtropStereoValue.UNKNOWNmeans that the angle is near 0 or Pi (5° threshold), or not defined (2d structure without wedge information). -
AtropStereoValue.WIGGLYmeans that either the axis, or any of the bonds in the rings connected to the axis is WIGGLY.
Axial stereo descriptor
If two stereoactive atoms (atoms with at least three different ligands) are connected by an even numbered chain of rigid parts then axial stereo information can be defined on the ligands of the stereactive atoms. These ligands are the ones which are not in the chain of the rigid part.
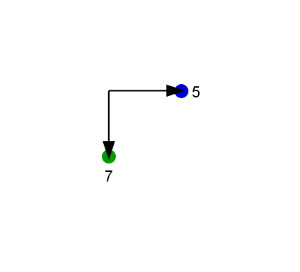
Here we consider the chain of rigid parts as an axis. Number the ligands at the end of the chain according to their atom indices. Implicit Hydrogen has the lowest number, -1.
 |
 |
|---|
The angle between two bonds connecting the highest indexed ligands on both sides of the axis is considered. On the example these are the bond between atoms 5 (marked blue) and 1 (one end of the axis) and the bond between atoms 7 (marked green) and 3 (other end of the axis).
-
AxialStereoValue.EVENmeans that the angle is positive (between 0 and Pi). -
AxialStereoValue.ODDmeans that the angle is negative (0 and -Pi). -
AxialStereoValue.UNKNOWNmeans that the angle is near 0 or Pi (5° threshold), or not defined (2d structure without wedge information). -
AxialStereoValue.WIGGLYmeans that any of the non-rigid-part-bonds connected to any of the stereoactive atoms is WIGGLY or there is a CIS/TRANS double bond on the chain of rigid parts between the stereo active atoms.
CIS/TRANS stereo descriptor
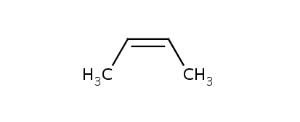
In general, single bonds are rotatable, but double bonds are not. If the substituents on each side of the double bond are different then two diastereomers of the molecule can be distinguished based on the orientation of the ligands. Two substituents located on the same side of the reference plane defined by the double bond are referred to as cis isomer, otherwise, if the two substituents are located on the opposite side of the reference plane then it is referred to as trans isomer.
 |
 |
||
|---|---|---|---|
CisTransStereoValue.TRANS |
CisTransStereoValue.CIS |
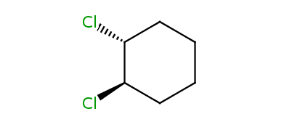
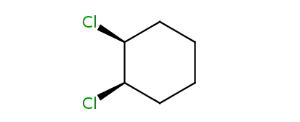
Alicyclic compounds can also display cis-trans isomerism. In this case the so called rigid part becomes non-rotatable due to the constrain of a cycle.
 |
 |
|---|---|
| trans -1,2-dichlorocyclohexane | cis -1,2-dichlorocyclohexane |
-
CisTransStereoValue.CISmeans that the ligands with highest indexes are in CIS position. -
CisTransStereoValue.TRANSmeans that the ligands with highest indexes are in TRANS position. -
CisTransStereoValue.WIGGLYmeans that any of the non-rigid-part-bonds connected to any of the stereoactive atoms is WIGGLY or there is a CIS/TRANS double bond on the chain of rigid parts between the stereo active atoms. -
CisTransStereoValue.UNKNOWNmeans that CIS/TRANS value can not be defined, e.g. there is a ligand which is collinear with the rigid part to which it is attached (with 5° threshold), or ligands at one end are not in opposite directions of the axis, or wedge information on bonds in 2D is not consistent.
Examples of unknown CIS/TRANS situations:
-
Same wedge bond at one of the stereoactive parts:

-
Wedge bond is only at one of the stereoactive parts:
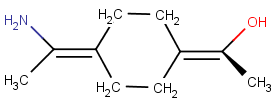
-
No wedge bond, one of the ligands is collinear with the axis:
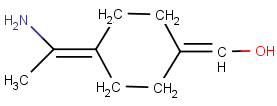
-
No wedge bond, ligands are on the same side of the axis:
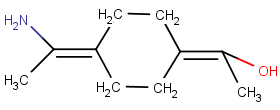
Rigid Parts
Stereo active atoms are considered at the ends of rigid parts or chains of rigid parts. The following substructures are considered as rigid parts:
-
Double bonds.
-
Four or six membered rings.
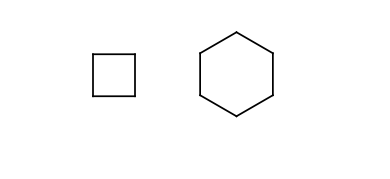
-
Four-four and six-six membered fused ring pairs.

-
Odd membered rings having lower than eight members, connected to each other directly or connected by intermediate four or six membered rings. Connection of two rings means that they share exactly one common bond (all rings are fused). The intermediate even membered rings have to connect to other rings by bonds on the opposite sides.
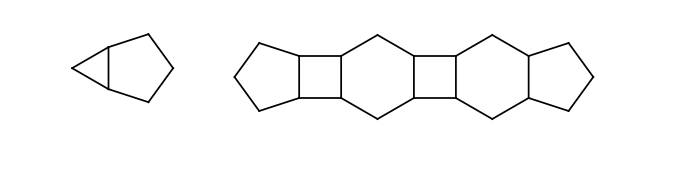
The following chain of rings is not a rigid part because the second intermediate ring connects to its neighbors not by opposite bonds (marked green):

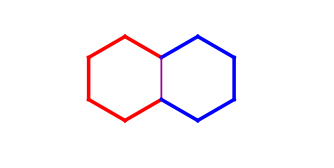
The rigid part is represented by the atoms and bonds of the outer hull. The outer hull is detected as the symmetrical difference of all ring bonds contained in the system. The hull of a rigid part is marked green on the following example. There is always an opposite atom for each contained atom in a rigid part, e.g. atom 13 is the opposite of atom 17 on the example.

The same ring can be contained in more than one rigid system (e.g. all six-six membered ring pairs are detected as two separate six membered rings as well). On the example below the red ring and the blue ring are detected as rigid parts (their common bond is purple) as well as the union of the two fused rings (marked with thicker bonds).
Not supported rigid parts
The more complex cases listed below are also rigid ring systems which are not supported yet, these will be handled in future releases:
- Condensed ring systems consisting of more than two fused rings.
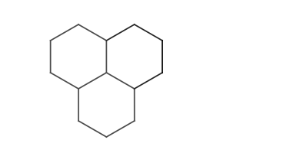
- Molecules where ring chains are fused (where at least three rings share the same common bond and this is their only interesection). In this case it is possible that not all chains will be found. The common bond between atoms 4 and 10 shared by chains of rings between the three odd membered rings at the ends is marked red on the example below.

Chain of Rigid Parts
Stereo active atoms can be connected to each other by a chain of rigid parts where the rigid parts are connected to another by exactly one atom (which is a spiro join for rings) and these connections are on opposite atoms of the rigid parts. On the example below each rigid part defining the chain of rigid parts between atoms 12 and 33 is marked with different color.
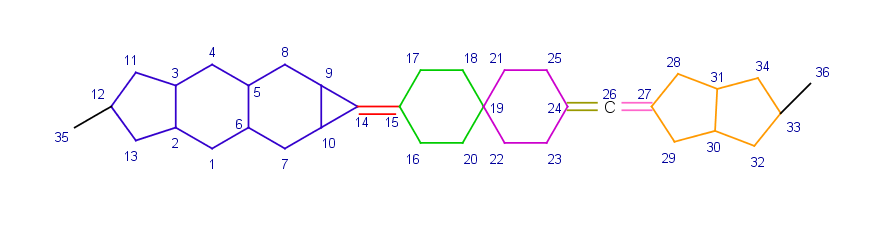
The type of the stereo active descriptor depends on the number of rigid parts in the chain of rigid parts: it is axial if the number is even, cis/trans if the number is odd. On the above example the number of rigid parts between atoms 12 and 33 is seven thus their ligands with indexes 35 and 36 are in trans position.
Stereo Active Part (SAP)
Stereo active part of a molecule is an atom with 0 to 4 ligands that can have tetrahedral stereo information, or with another stereo active part CIS/TRANS, ATROP or AXIAL stereo information.
Stereo centers with more than 4 connections are not supported yet.
Programmatically, SAP consists of an integer that is the index of the stereo center atom and an array of integers listing the indexes of the ligands of the stereo center atom. If the molecule contains S-groups, these atom indexes are referring to the expanded molecule.
CIPStereoDescriptor types
TetrahedralStereoDescriptor
The stereo active center and its ligands are defined in one SAP. The TetrahedralStereoValue defines the spatial arrangement of the ligands around the stereocenter (UNKNOWN, ODD, EVEN, WIGGLY).
AtropStereoDescriptor
The stereo active single bond (with hindered rotation) which connects two aromatic rings is represented with two SAPs. Each StereoActivePart contains a stereo active atom which is the endpoint of the single bond and the ligands which are connected by aromatic bond to this active atom. (Such a way the stereo active single bond ligand is not part of the ligands array.) The AtropStereoValue defines the spatial arrangement of the ligands around the stereo active single bond (UNKNOWN, ODD, EVEN, WIGGLY)
AxialStereoDescriptor
The two stereo active atoms (atoms with at least three different ligands) which are connected by even number of rigid parts are represented with two SAPs. Each StereoActivePart contains the stereo active atom and the ligands of the stereactive atoms. These ligands are the ones which are not in the chain of the rigid part.. The AxialStereoValue defines the spatial arrangement of the ligands around the two stereo active atoms (UNKNOWN, ODD, EVEN, WIGGLY)
CisTransStereoDescriptor
The two stereo active atoms (atoms with at least three different ligands) which are connected by odd number of rigid parts are represented with two SAPs. Each StereoActivePart contains the stereo active atom and the ligands of the stereactive atoms. These ligands are the ones which are not in the chain of the rigid part.. The CisTransStereoValue defines the spatial arrangement of the maximal indexed ligands around the two stereo active atoms (UNKNOWN, CIS, TRANS, WIGGLY)
API usage example
The following example demonstrates how to use the CIPStereoRecognizer and the equals/equalsTo methods. Under the code, you can find the two molecules (m1 and m2). Example for CIPStereoRecognizer
//read or build the molecules
Molecule m1;
Molecule m2;
/* ... */
int[] map = { 2, 1, 3, 0 };
Collection<CIPStereoDescriptor> descriptors_m1 = CIPStereoRecognizer.calculateCIPStereoDescriptors(m1);
Collection<CIPStereoDescriptor> descriptors_m2 = CIPStereoRecognizer.calculateCIPStereoDescriptors(m2);
CIPStereoDescriptorIface desc1 = (CIPStereoDescriptorIface) descriptorArray_m1.toArray()[0];
CIPStereoDescriptorIface desc2 = (CIPStereoDescriptorIface) descriptorArray_m2.toArray()[0];
System.out.println(desc1.equals(desc2)); //false
System.out.println(desc1.equalsTo(desc2, map)); //true |
 |
|---|