Bulk Upload
Upload compounds
Supported formats
The Upload page is designed to simultaneously handle multiple submissions imported from an SD file according to configurable system settings.
The following molecule formats are supported: SDF, SMILES, SMARTS, MRV, CSV.
{info} In the case of SDF files the structure field is mandatory. Even if the structure part contains an empty structure it should be still mapped, otherwise, the upload will fail.
The structure within the CSV file can be in ChemAxon Extended SMARTS (1), SMARTS (2), ChemAxon Extended SMILES (3), or SMILES (4) format.

{info} In the CSV file, the header is mandatory, columns can be comma or semicolon separated. The first column must be the the structure column followed by any other fields.
{info} The structure column can contain a valid chemical file format as listed above for registering Single structure, or can be left empty for registering No-structures. At least one field (e.g. Id or additional data) must be mapped prior to initiating the upload process or the upload will fail.
{primary} Note that in case of CSV files the system might not automatically recognize the fields, so manual mapping should be made in order to successfully upload the file.
Uploading an SDF that contains compounds with salts
Stripping salts/solvates from compounds is performed when the "Analyze Salt Solvate Fragments" Source dependent Registration option is ON. Only salts/solvates that are present in the Salts &Solvates Dictionary can be stripped.
Uploading an SDF that contains compounds and salts Ids
During the upload, the user can provide the salt/solvate Ids and multiplicities for each compound structures if needed. Salts/solvates from the Salts &Solvates Dictionary can be referred by their Id and multiplicity in the SDF within a data field that is going to be mapped with the "Version salt/solvate ID" and "Version salt/solvate multiplicity" fields (Figure 1)
Uploading multi-component compounds
Please note, that the Bulk Upload does NOT support the registration of multi-component compounds. In the case when the loaded SDF file contains multiple components within one structure, this will be registered as a "single" compound consisting of more components (not as a Mixture, Formulation or Alternate). Or, as a solution, a multi-component checker can be introduced, in which case these records will not be autoregistered but will fall to the Staging area, from where these can be manually registered as multi-component compounds.
Invalid structures
Unreadable f ile format : When a molfile cannot be parsed and no information can be extracted from it (because it might be corrupted or does not follow the expected file format specification).
Inconsistent f ile format : Even though a valid molfile (can be parsed according to syntax specification, passes format validation) is present, it can happen that the molecule described by the file does not make any sense and is likely wrong.
Unsupported file format : ChemAxon may not know that a section of the file contains structure-relevant information and simply skip parsing that information. Often caused by modifications to the file format specifications introduced by vendors or organizations, that are not widely used and not sufficiently described publicly. In other words: we might miss extracting data, that is not supposed to exist according to the publicly available file format specs and we cannot assume what type of data and what its context might be.
Using Compound Registration invalid structures can be uploaded successfully. The compounds will be stored as "No structures" while the original structure as the text will be stored as additional data. Later the "No structures" can be amended on the parent compound level and the correct structure can be provided (the Ids will be kept).
In order to prepare the system for storing the original (invalid) molfile as a text, an additional data field should be created (Administration/Forms and Fields) that has the "errorStructure" as field identifier.
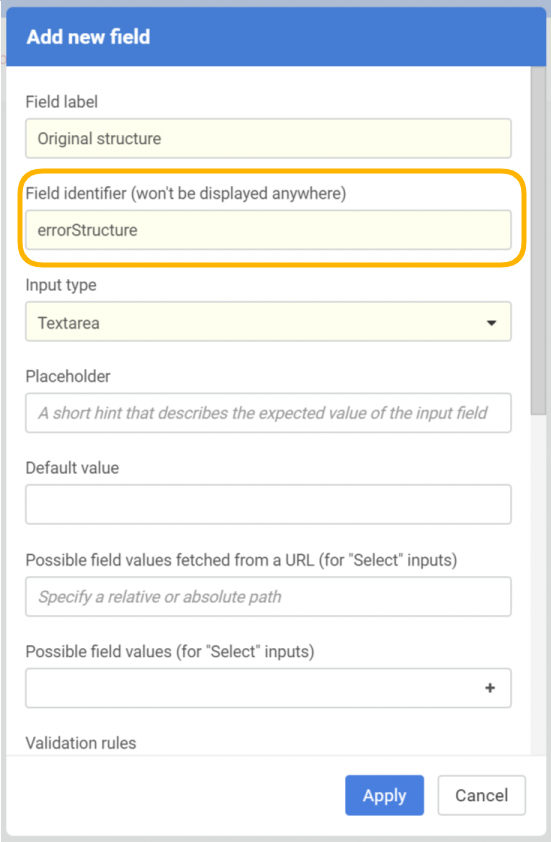 |
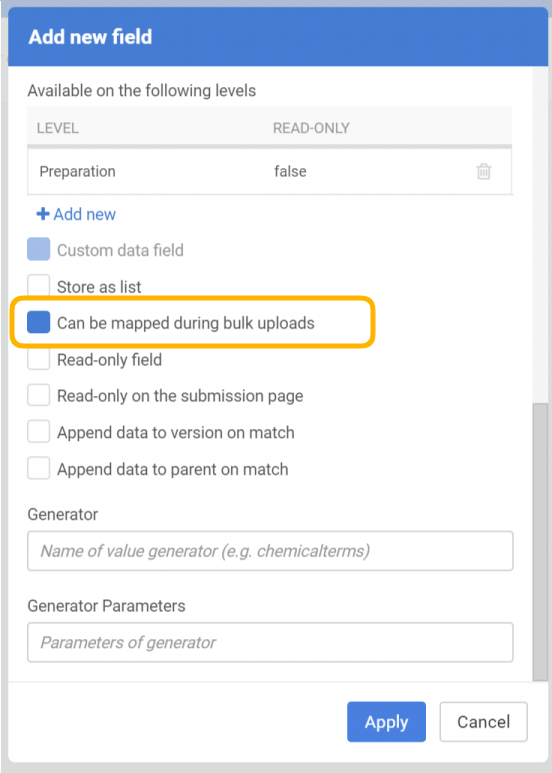 |
|---|---|
| New field configuration |
On the Upload page the previously created additional data field can be appended:
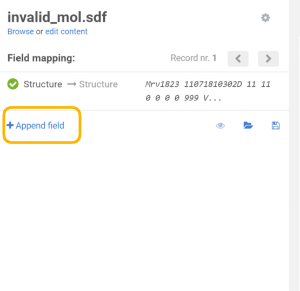 |
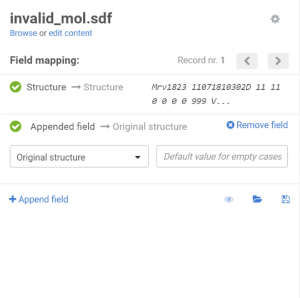 |
|---|---|
| Appending an additional field |
After uploading the file, the records that have invalid structures will be converted to "No structures" and will have the structure as text in a field stored as additional data to the compound.
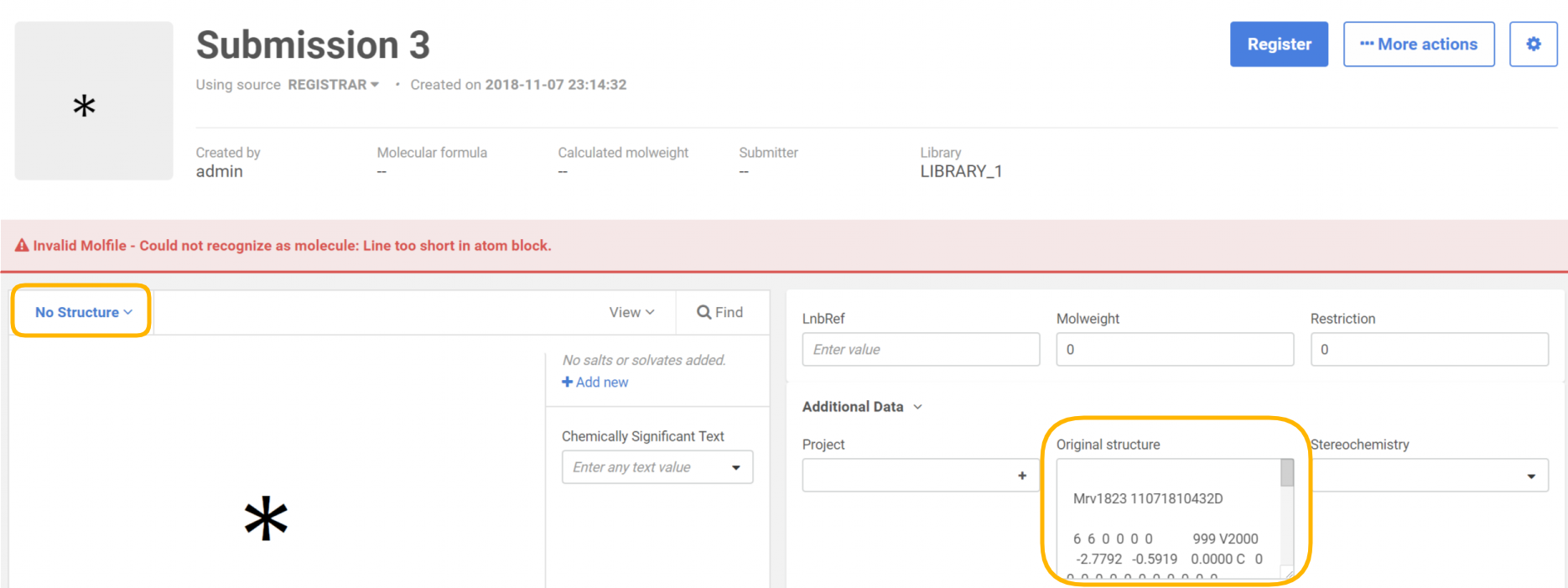 |
|---|
| During registration on the Staging area/Submission page |
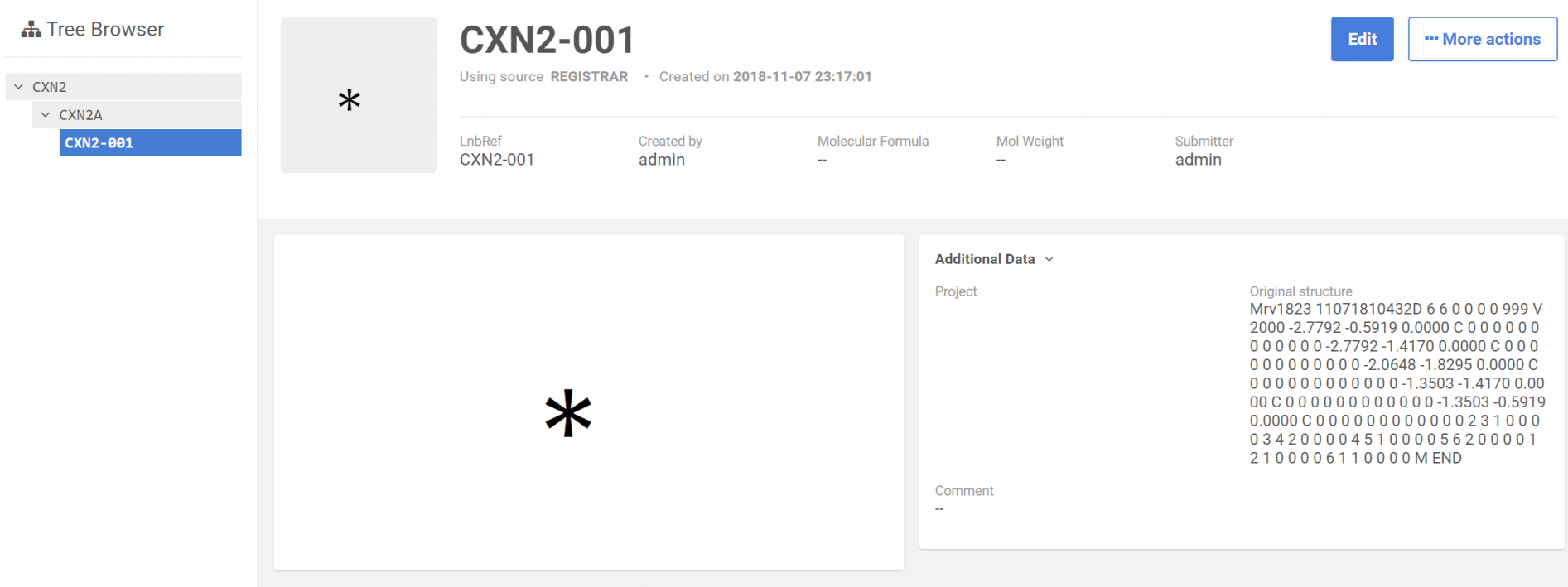 |
|---|
| After registration: Browse page |
Submissions can be registered one by one from the Staging area (Submission page) or can be registered through a bulk operation:
Register all from the Upload summary page
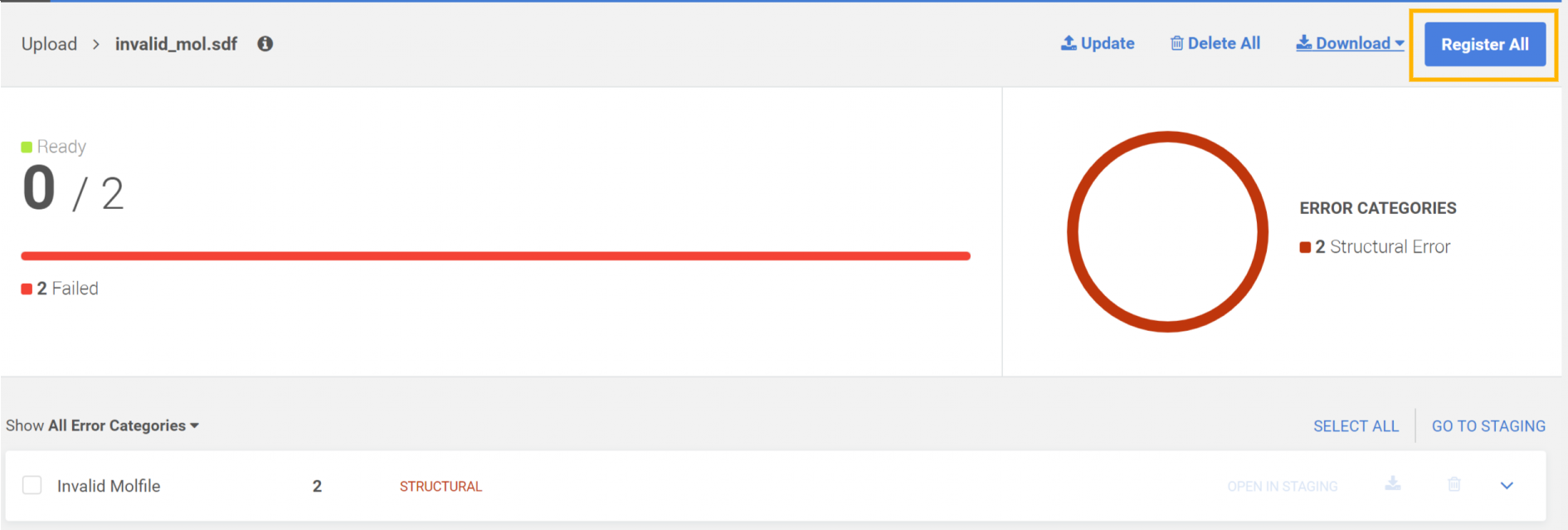 |
|---|
| Registering from the Upload summary page as a bulk operation |
Register all from the Staging area
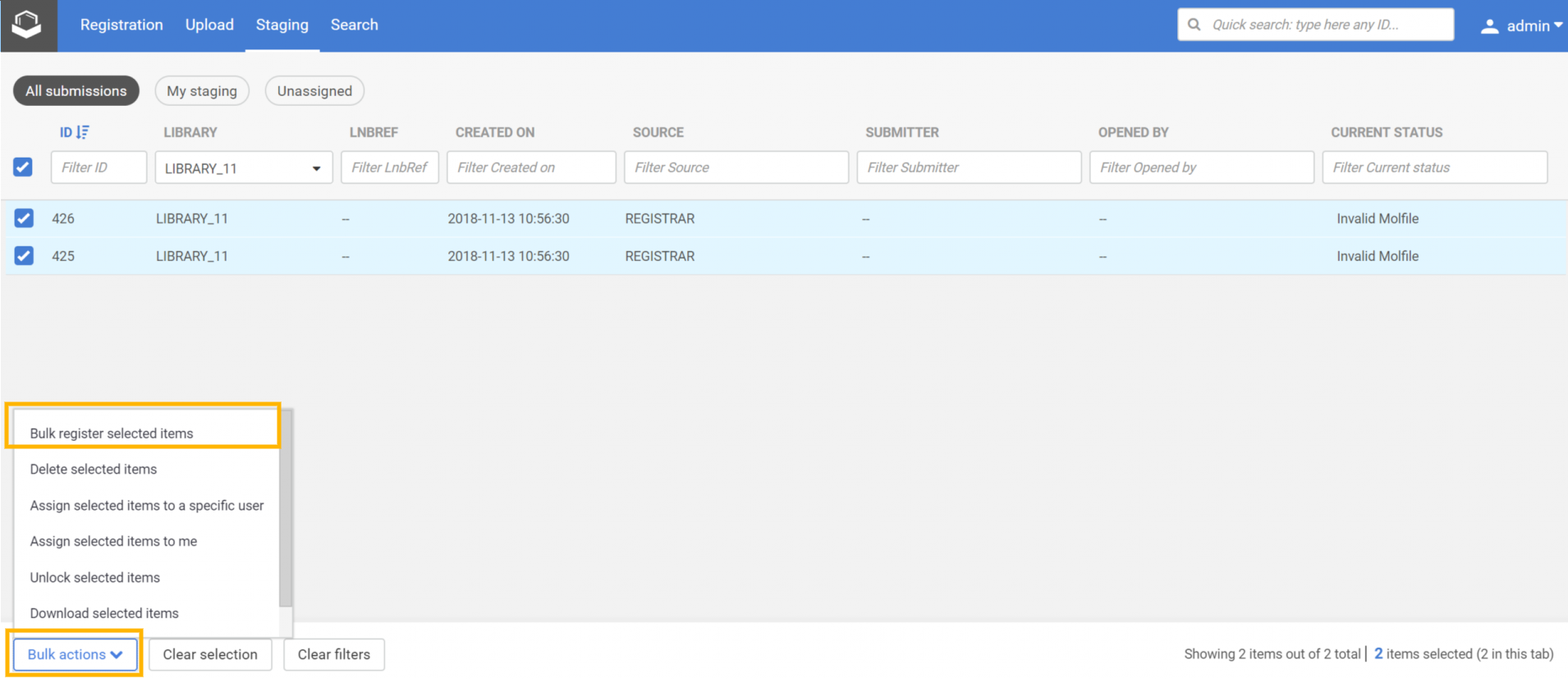 |
|---|
| Step 1: Select the submission and choose the Bulk register selected items option |
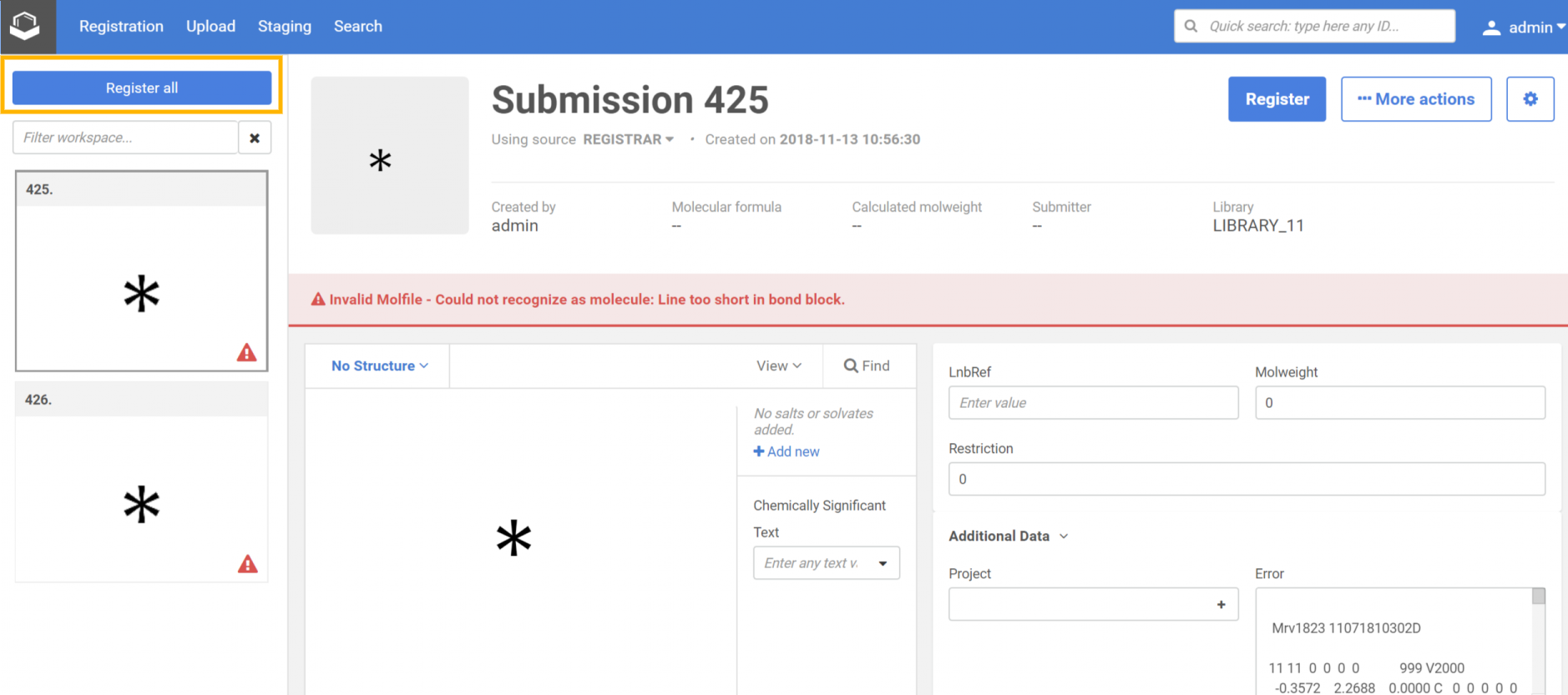 |
| Step 2: On the Submission workspace click on the Register All |
Upload page
The Upload page consists of two main sections, the File uploader and the All uploads.
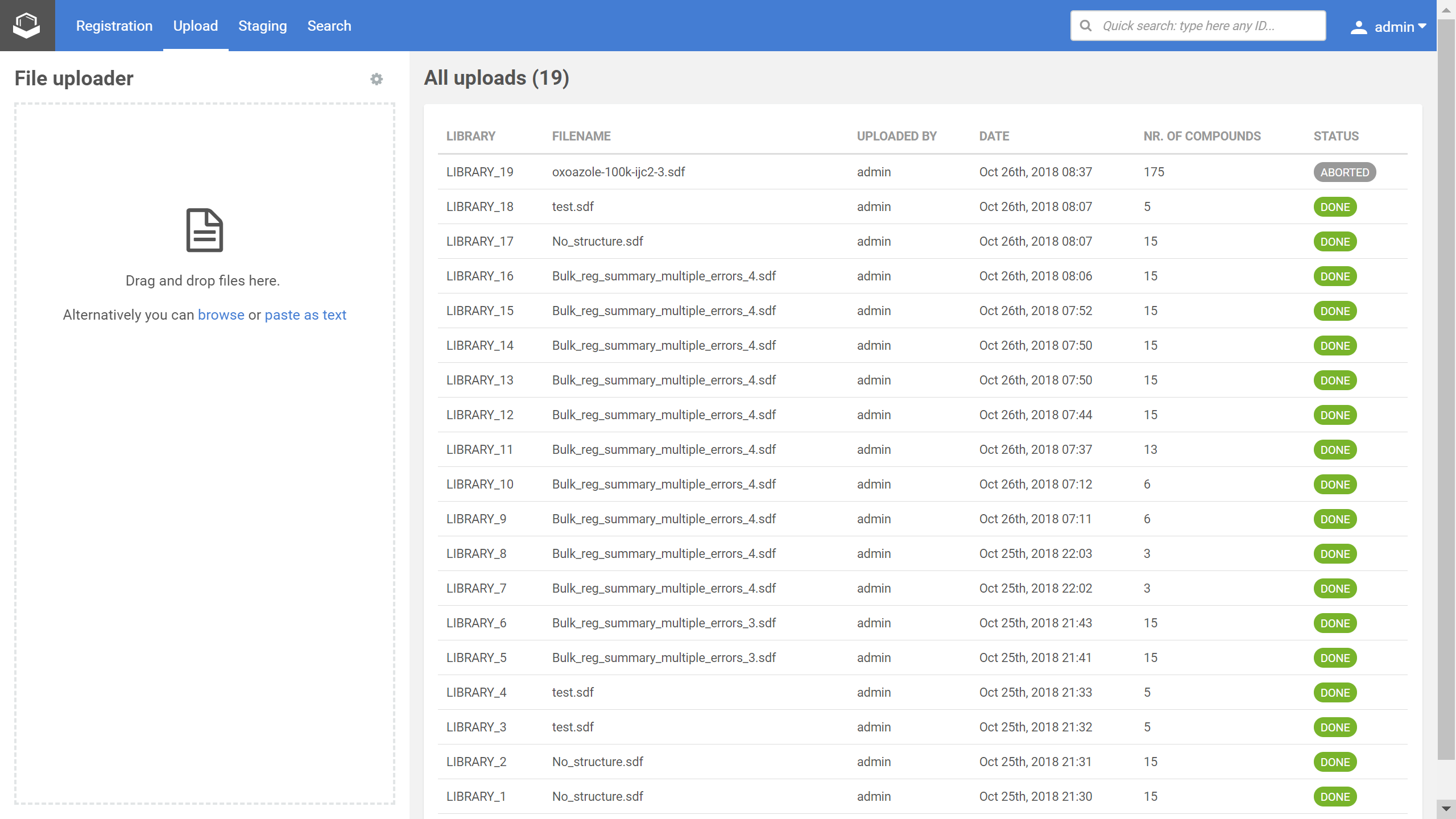 |
|---|
| Figure 1. The Upload page with the recent uploads |
An SD file can be dragged and dropped into the File uploader, but you can also browse for files or paste as texts.
After the file to be registered has been selected, the user is allowed to navigate with the aid of arrows between the first records, while the accompanying data fields are displayed. The available data fields are determined based on the first ≈250 records. If more than 250 records are present, the [Scan more] button will be available. Each click on the [Scan more] button will process 250 more structures. If no more structures are available for examination, the button is not displayed.
{info} While the Scan more button is active the user is always notified about the number of compounds processed from the SDF
When uploading compounds, the data accompanying the structures can be e.g. LnbRef, Lot ID, PCN, CN, CST, Submitter, Salt ID, Salt multiplicity, etc. While the external ID fields (according to the actual business rules, e.g. LnbRef, Lot ID) can be configured to be mandatory or not for the registration, the structure (that can be an empty structure) is usually necessary to be able to initiate a registration (exception "No structures" in case of CSV files). Each column/field of the SD file can be mapped to any of the mandatory and optional fields of the structure object. The mapping can be set manually by selecting the appropriate parameter from the drop-down list above each column. E.g. "user" from the SD file can be mapped with "Lot submitter" from the DB table. The mapping can be saved in a text file using the [Save mapping] button and can be used again for other bulk upload registrations using the [Load mapping] button. When a field exists but contains a value just for some structures, there is a possibility to set a default value for the empty cases. After mapping the fields and setting a default value for the empty cases, the existing values will not be lost, just a value for the empty ones will be set.
It is possible to set specific Ids (PCN and/or CN) for the registered compounds, even during a bulkload process. For this, you just need to map the PCN and/or CN fields to the corresponding columns of the SDF file.
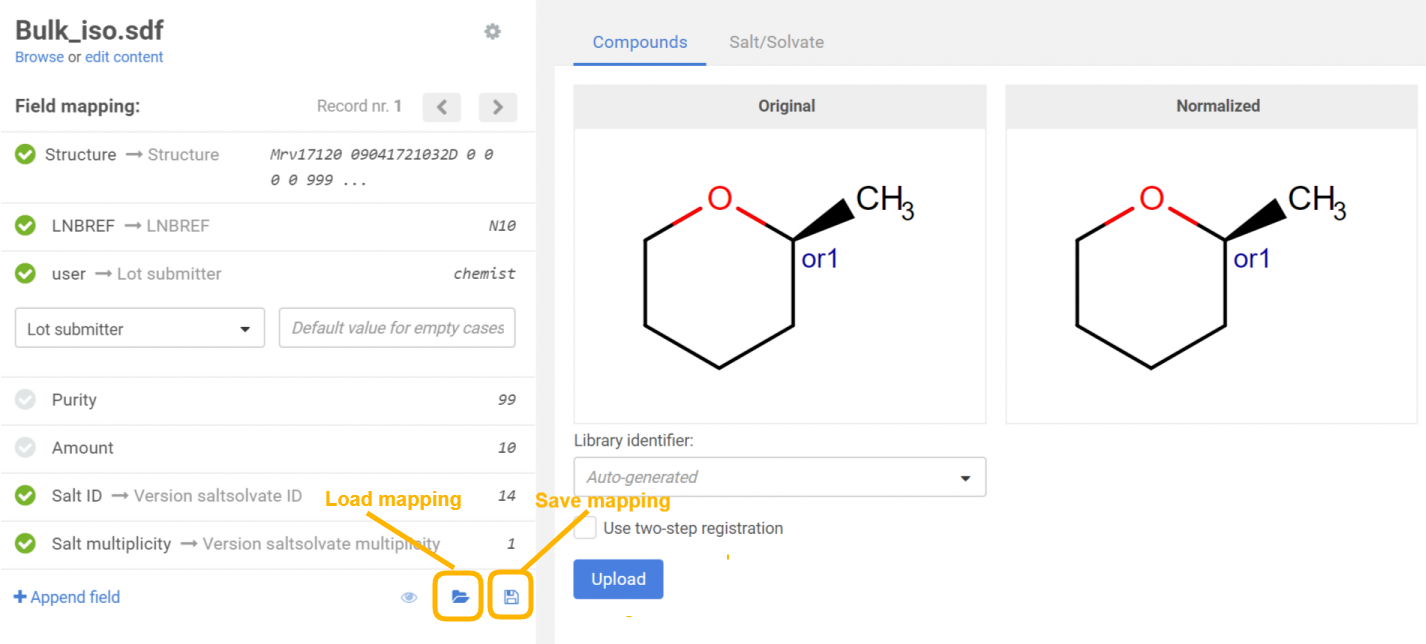 |
|---|
| Figure 2. Field mapping before uploading a file |
For a bulk upload process, the source can be set. According to the selected source, Checkers (and fixers) and Registration options become available, that will be applied for all the structures of the selected SD file.
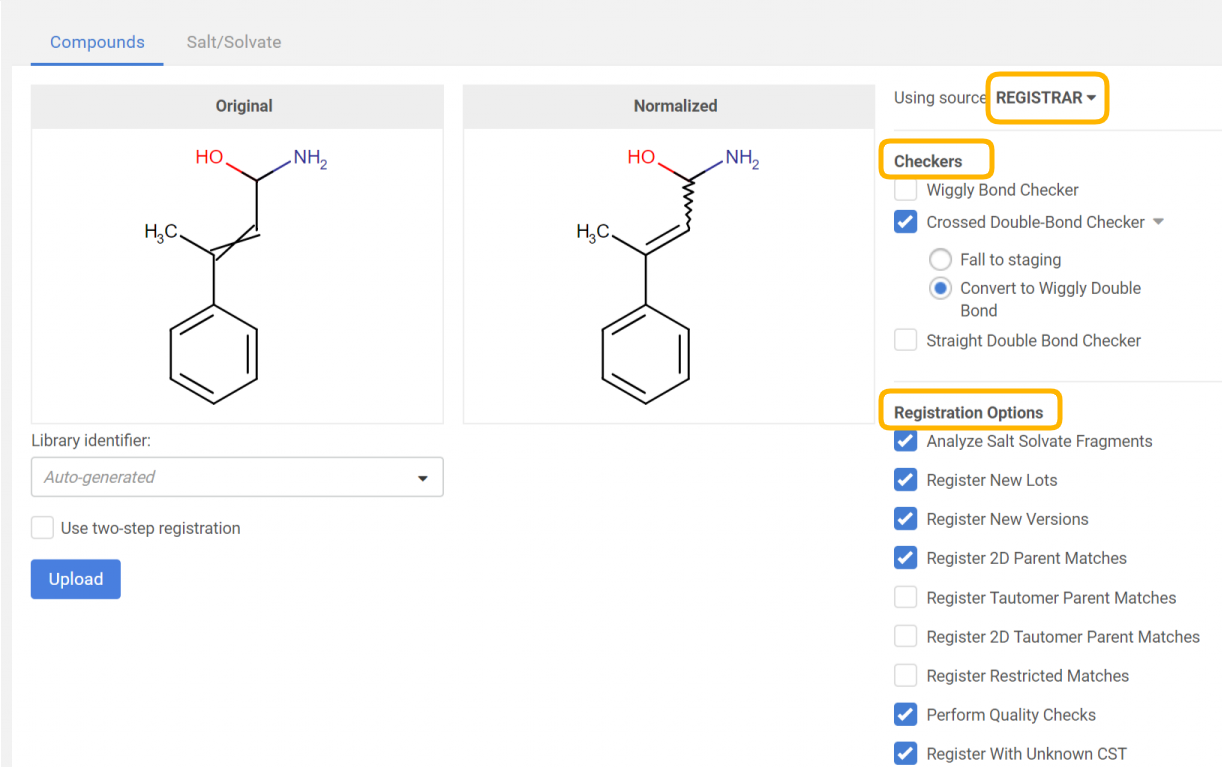 |
|---|
| Figure 3. Applying a structure checker and fixer. The original and normalized structures are displayed. |
By clicking on the [Upload] button, the registration process will run according to the autoregistration process, with the only difference, that here the selected structure fixer options will be applied to the structure before the actual registration and the "normalized" strructure can be compared with the "original" one (Figure 3).
Two-step registration
The two-step registration option allows the user to pre-register the compounds to the staging area in order to check them before registration.
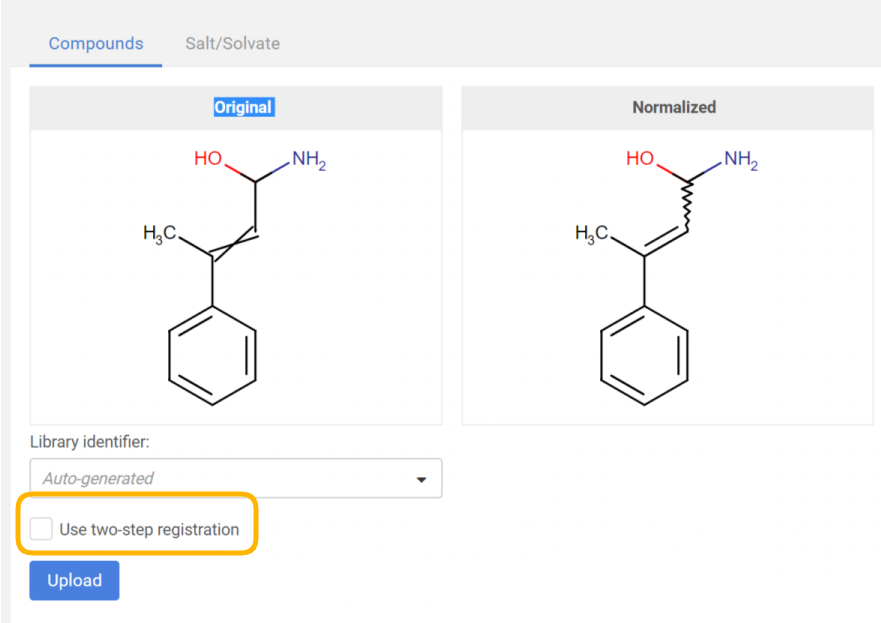 |
|---|
| Figure 4: Two-step registration option on the Upload page |
During the two-step bulk upload, the user is directed to the pre-registration summary page, where the report of the process can be seen (Figure 4). The submissions that would be registered fine, should end up in the Staging area and also appear in the summary page with "Ready for registration" status. Besides the submissions with "OK" status the failed submissions are also listed, similarly like in the case of a regular bulk upload process (Figure 5).
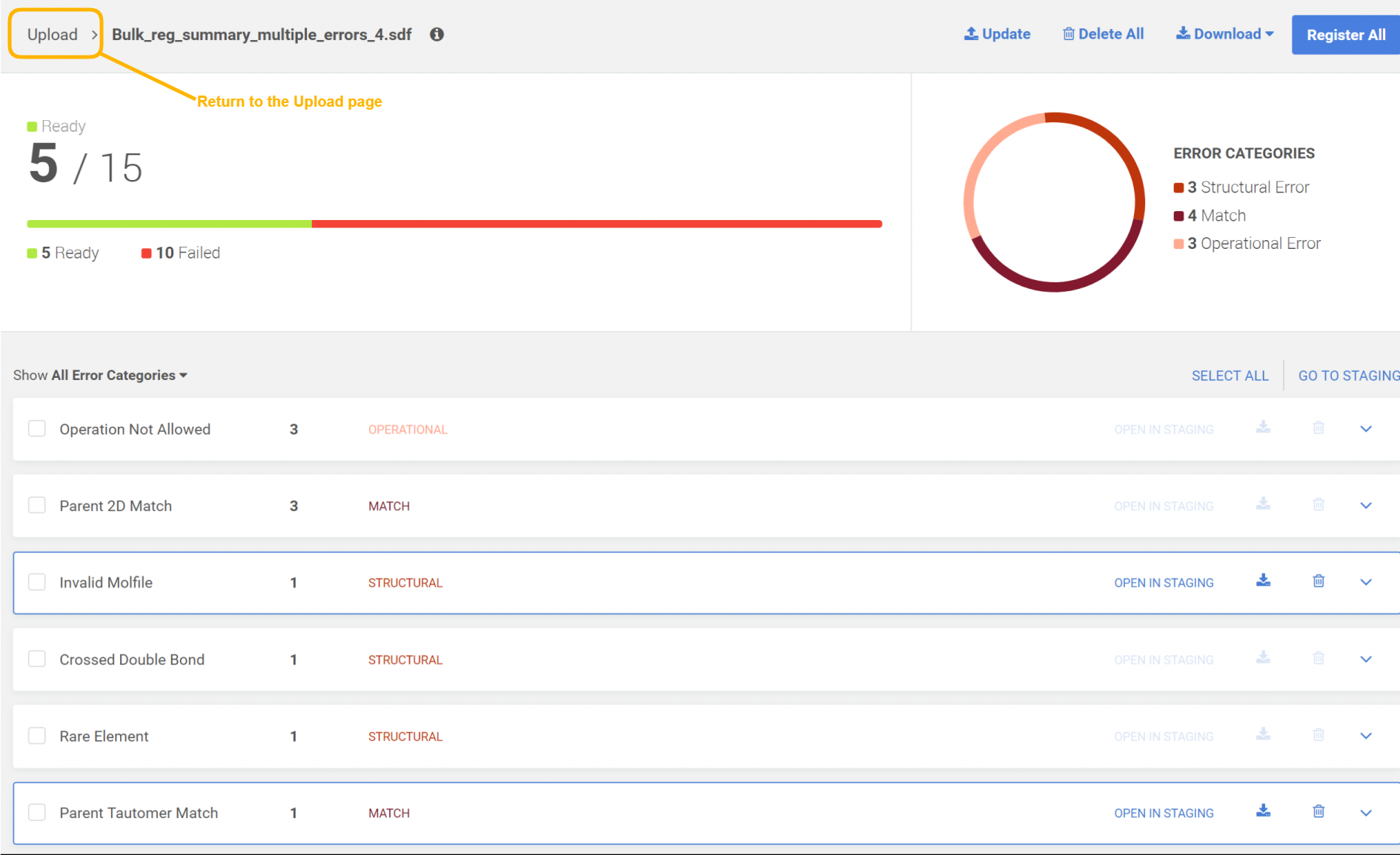 |
|---|
| Figure 5. The bulk upload summary page |
Bulk upload summary
By clicking the arrow (Figure 5, next to the file name) the user will return to the Upload page where all the uploads are listed with the information of each upload (Figure 1). By clicking on a certain row on the Uploads list the user has navigated again to the Bulk upload summary page.
By clicking on the info sign more details of the given bulk upload process are shown.
The user can easily download one or all the problematic structures and fixed them locally and then re-upload them with the update option, and register the whole uploaded set without errors. The update process performed based on matching submission IDs, in case of non-matching submission ID, the update attempt will be ignored. When the update is finished the number of successful and ignored structure updates appear.
The bulk upload process is also visible on the Dashboard page, where a process indicator can be seen and the upload process can be also canceled. When the upload is finished, we are informed about it in the Bulk Uploads section. The uploaded submission are found either in the Recently Failed or in Recent Successful Submissions section of the Dashboard page. A click on a submission on the Recently Failed section will open the submission on the Submission correction page and if not registered, the submission will be moved from the Registered by me table to the Assigned to me table. Submission from the Recent Successful Submissions section is opened on the Details page. On the Dashboard only the user's own uploads are visible but on the Upload page with the SUPERVISE_UPLOAD role, the user can see other uploads initiated by other users.
Display the successfully registered compounds
In case when a regular bulk upload process is made the successfully registered compounds appear in the Registered category.
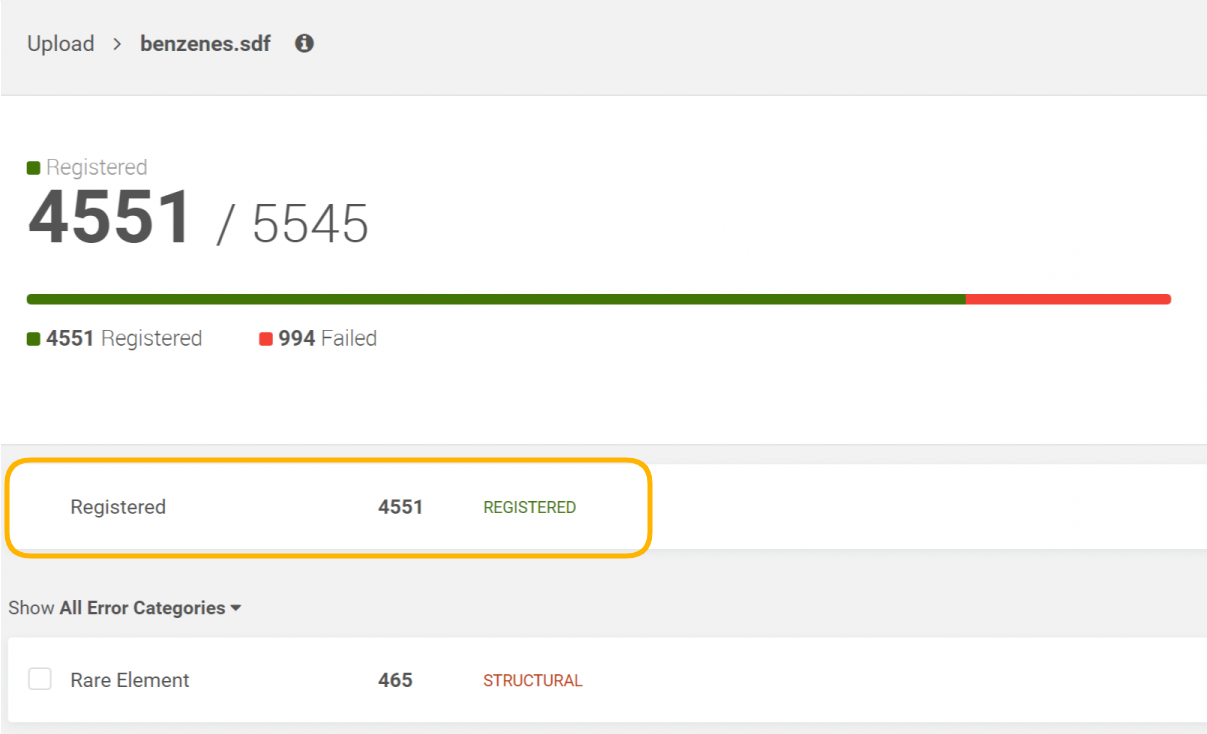 |
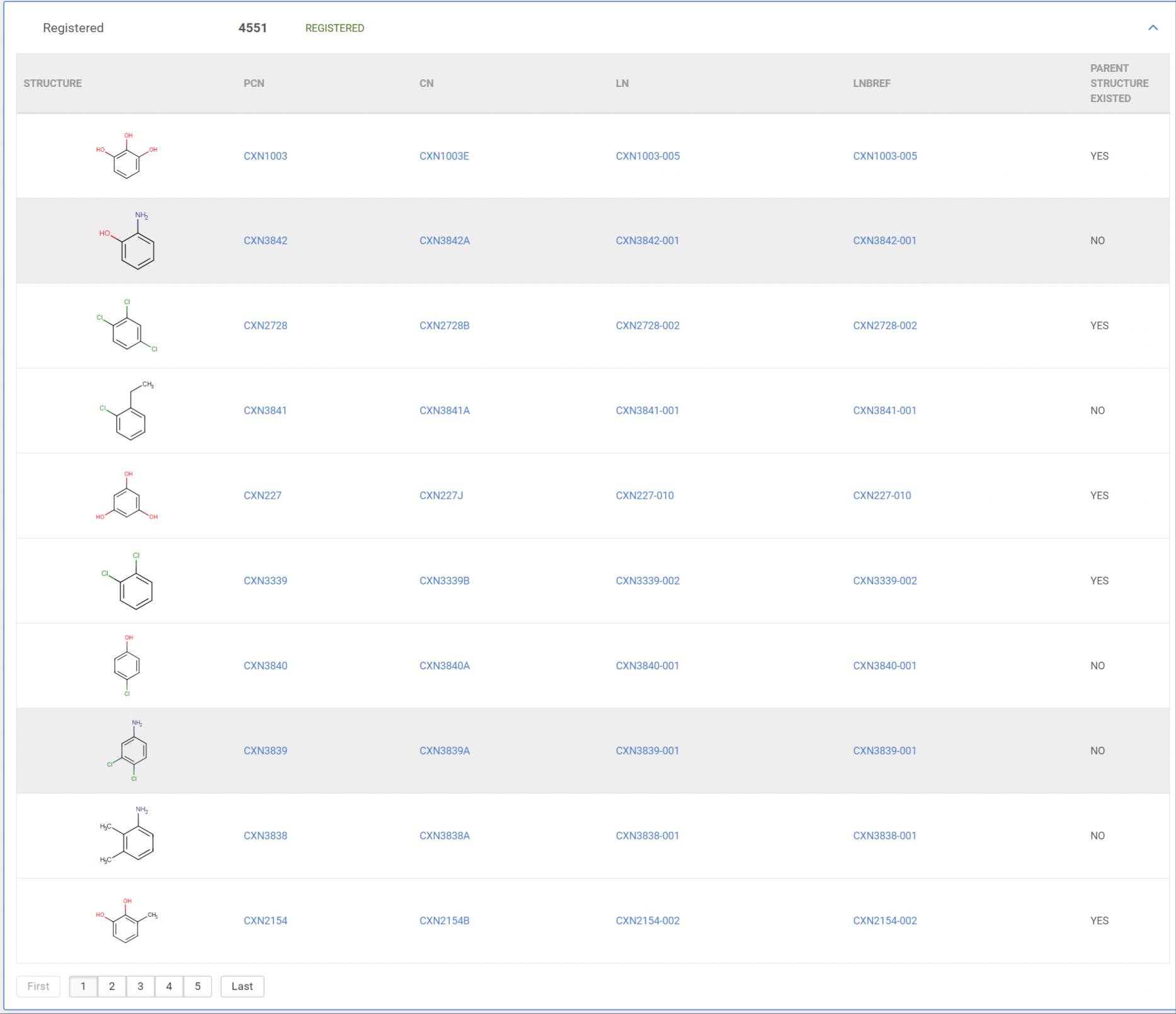 |
|---|---|
| Figure 6. Registered compounds |
The list of successfully registered compounds is paginated and contains all relevant Ids (PCN, LN, LnbRef, and CN-configurable) and the info whether the compound was a newly registered one or not.
Display the failed submissions by error type
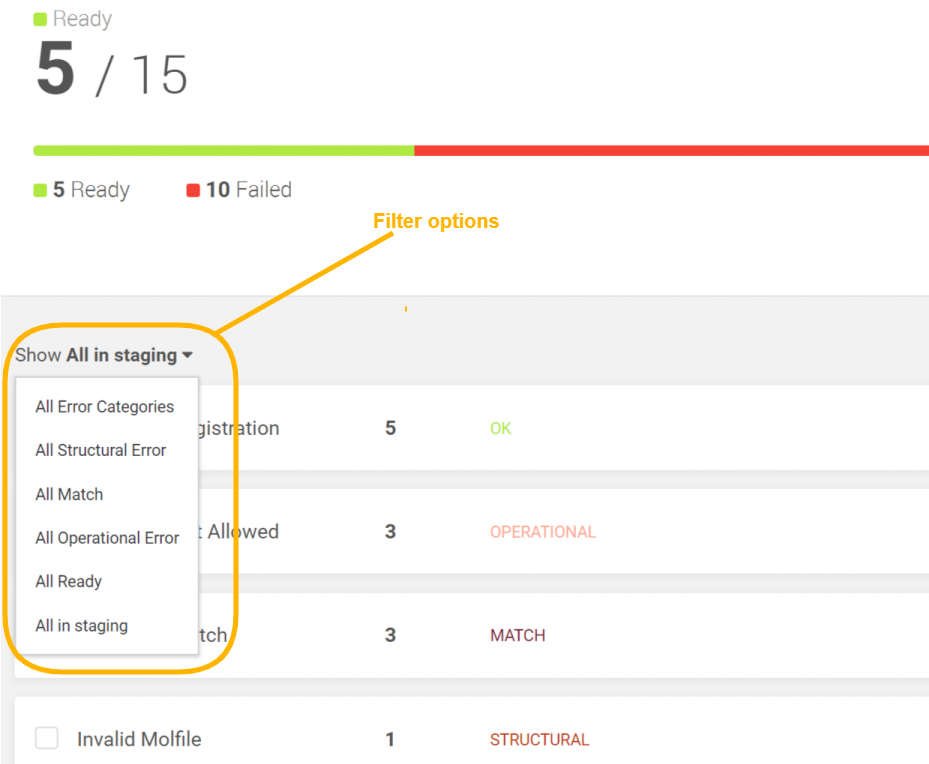
The erroneous submissions are categorized by the following types: operational, structural and match. A donut chart (Figure 5) represents the submission distributions according to the error type.
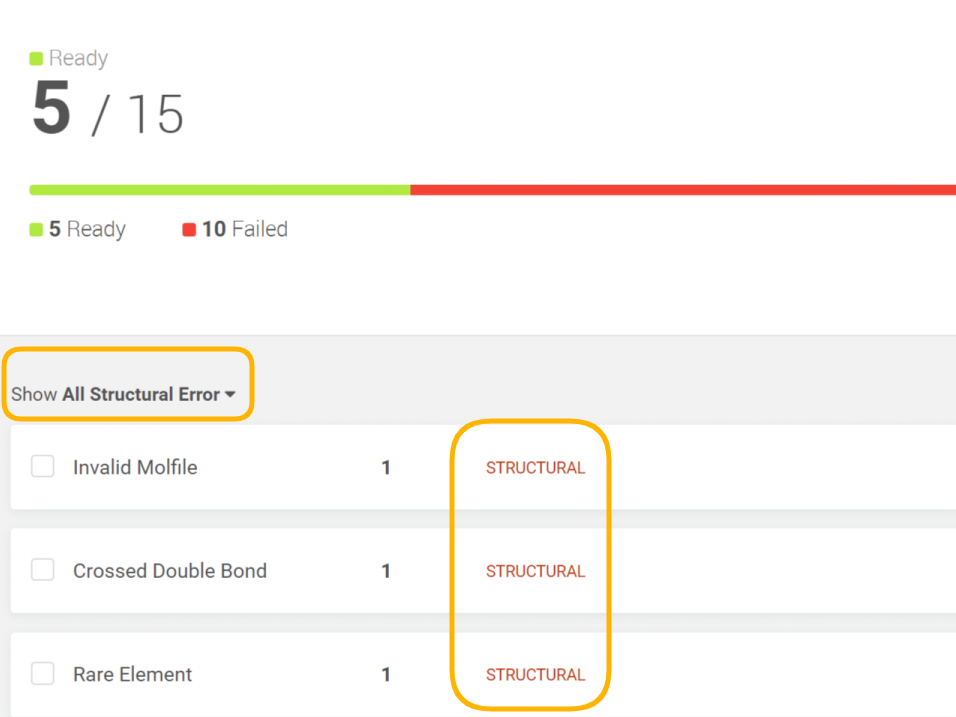
The user can filter out the submissions by error type (Figure 7).
 |
 |
|---|---|
 |
|
| Figure 7. Filter options on the Pre-registration summary page |
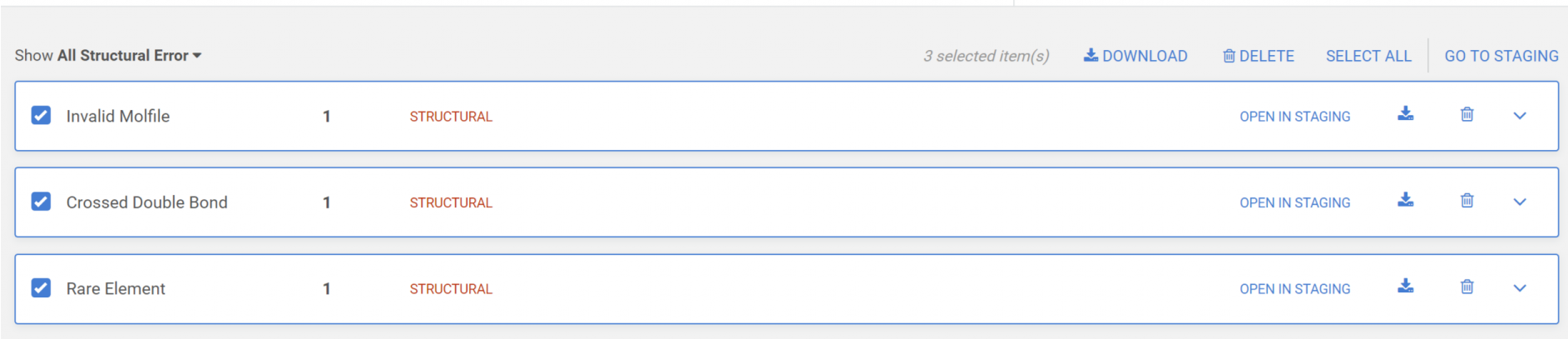
Submissions can be downloaded, deleted or opened in the Staging area for further analysis.
Download submissions
All submissions or just the selected one by error type can be downloaded in SDF format.
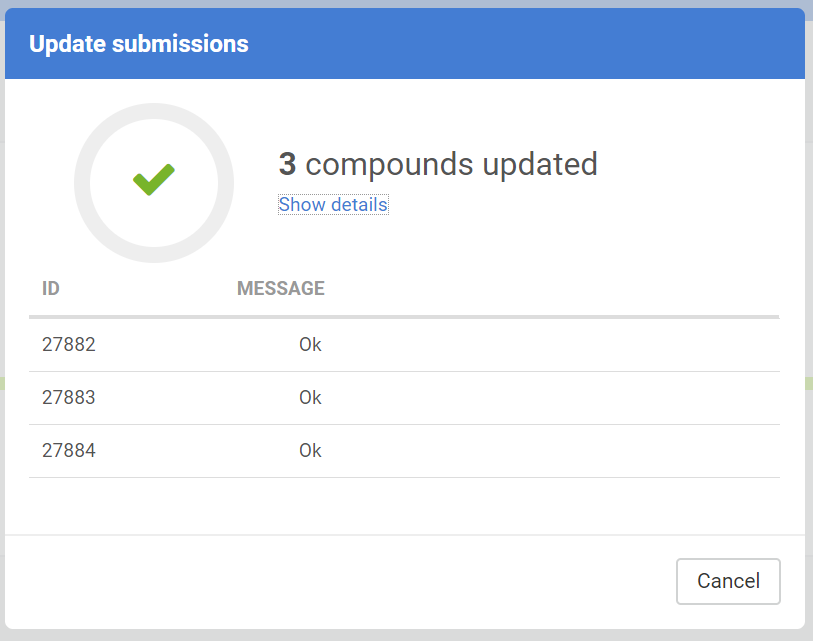
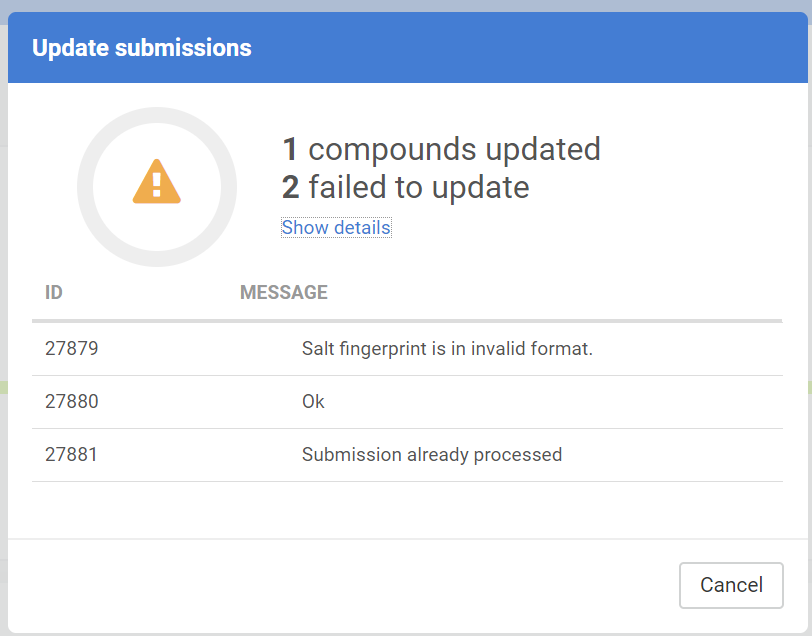
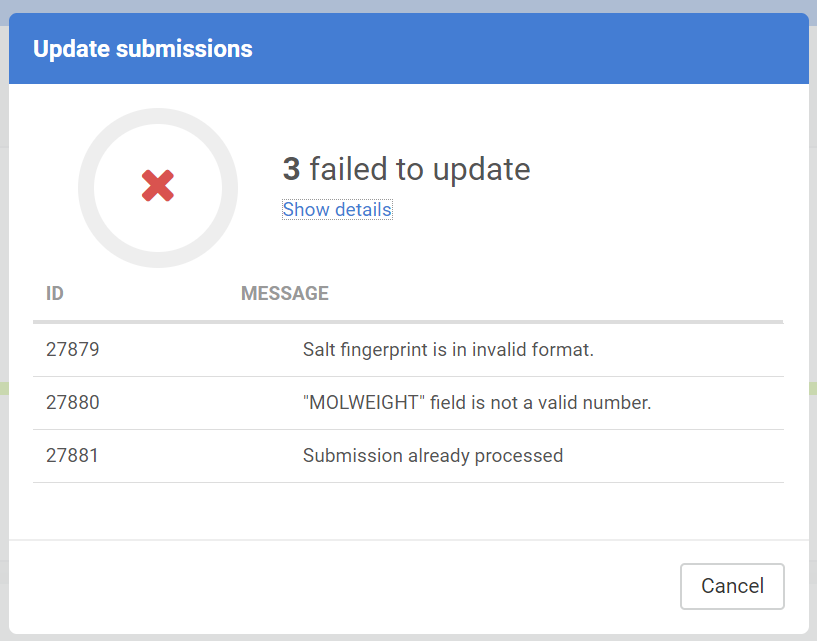
Update submissions
The downloaded submissions can be corrected outside the Compound Registration system and can be later re-uploaded in an update process. The update process can be either performed in the Staging area or on the Upload Summary page.
The submission update is not always successful, the Update submission report shows a summary of the update action:
 |
 |
 |
|---|---|---|
| Successful submission update | Partially successful submission update | Failed submission update |
The Update submission can have different messages:
| Message |
|---|
| OK |
| Submission already processed |
| Submission not found |
| No salt/solvate found with ID: ... |
| Salt fingerprint is in invalid format. |
| "MOLWEIGHT" field is not a valid number. |
| "Restricted" field is not a valid integer. |
| "Submission data is invalid. The source ... is unknown |
Download the upload report
The upload report in PDF contains all items from the uploaded SDF with all the relevant info. The registered structures appear in the first section of the PDF, the failed ones are after.
The registered structures section contains: REGISTERED, structure, PCN, CN (configurable), LN, Project, SaltSolvateComposition, Parent structure existed, MF.
NO additional data (besides Project) is displayed.
The failing submission section contains status, structure, submission Id, Project, SaltSolvateComposition, MF.
A PDF download confirmation window appears in case of large files: "You are about to export more than 1000 compounds, which may take longer. Are you sure you want to continue?"
Upload salt and/or solvates
Salts and/or solvates can be bulk uploaded to a salt dictionary in order to be used later as salt/solvate info next to the structures to be registered.
An SDF file containing salts and/or solvates can be uploaded in a similar way as the compounds. First, the file is browsed, then the Salt/solvate tab should be chosen.
 |
|---|
| Figure 8. Uploading salts and solvates |
In the Salt/solvate tab, a preview of the salt/solvate structure is available, and the type of the salts/solvates can be chosen. If the file contains only salts, then only salt name, if the file contains salts and solvates, then both salt and solvate name should be mapped to the proper fields from the SDF file. Otherwise "Missing mappings" message is received and the bulk load process cannot be continued. In order to process, after choosing the proper mapping fields, click on the [Upload] button.
In the case when some or all salts and/or solvates were registered, these will appear in the Successful imports table. The total number of successful imports is displayed and the table contains columns like Structure, Id, Name, and Type.
In the case when some or all salts and/or solvates failed to be registered, these will appear in the Failed import table. The total number of failed imports is displayed and the table contains columns like Structure, Name, Type, and Error. An error message can be e.g. "The name 'HCl' is already present in the database".
In order to be able to modify the salt/solvate structures and trying to load them later, the failed registrations can be downloaded in an SDF file using the [Download failed items] button located in the top right of the window. Once the downloaded SD file is modified it can be later again uploaded from the Upload menu of the Registration application.
On the Salt/solvate tab, after selecting the appropriate salt/solvate type, a "Force register" option is available. By using this option, on bulk upload, duplicate name or structures will be ignored.